class: center, middle, inverse, title-slide # Joining data from multiple sources ### Becky Tang ### 05.24.2021 --- layout: true <div class="my-footer"> <span> <a href="http://datasciencebox.org" target="_blank">datasciencebox.org</a> </span> </div> --- class: middle, center ## `mutate` (from May 20 lecture) --- ### `mutate` to add new variables .small[ ```r hotels %>% mutate(people = adults + babies + children) ``` ] --- ### "Save" when you `mutate` Most often when you define a new variable with `mutate` you'll also want to save the resulting data frame, often by writing over the original data frame. ```r hotels <- hotels %>% mutate(people = adults + babies + children) ``` --- ### Check before you move on ```r hotels %>% select(adults, children,babies, people) ``` ``` ## # A tibble: 119,390 x 4 ## adults children babies people ## <dbl> <dbl> <dbl> <dbl> ## 1 2 0 0 2 ## 2 2 0 0 2 ## 3 1 0 0 1 ## 4 1 0 0 1 ## 5 2 0 0 2 ## 6 2 0 0 2 ## 7 2 0 0 2 ## 8 2 0 0 2 ## 9 2 0 0 2 ## 10 2 0 0 2 ## # … with 119,380 more rows ``` --- class: middle, center # Working with multiple data frames --- ## Fisheries of the world Fisheries and Aquaculture Department of the Food and Agriculture Organization of the United Nations collects data on fisheries production of countries. <img src="img/05/fisheries-data.png" width="60%" style="display: block; margin: auto;" /> .center[ .hand[...] ] .footnote[ Source: https://en.wikipedia.org/wiki/Fishing_industry_by_country ] --- ## Load data ```r fisheries <- read_csv("data/fisheries.csv") ``` --- ## First look at the data ```r glimpse(fisheries) ``` ``` ## Rows: 216 ## Columns: 4 ## $ country <chr> "Afghanistan", "Albania", "Algeria", "American Samoa", "An… ## $ capture <dbl> 1000, 7886, 95000, 3047, 0, 486490, 3000, 755226, 3758, 14… ## $ aquaculture <dbl> 1200, 950, 1361, 20, 0, 655, 10, 3673, 16381, 0, 96847, 34… ## $ total <dbl> 2200, 8836, 96361, 3067, 0, 487145, 3010, 758899, 20139, 1… ``` --- ## Quick summaries of the data .xsmall[ ```r skim(fisheries) #skimr package ``` ``` ## ── Data Summary ──────────────────────── ## Values ## Name fisheries ## Number of rows 216 ## Number of columns 4 ## _______________________ ## Column type frequency: ## character 1 ## numeric 3 ## ________________________ ## Group variables None ## ## ── Variable type: character ──────────────────────────────────────────────────────────────────────────────────────────────────────────────── ## skim_variable n_missing complete_rate min max empty n_unique whitespace ## 1 country 0 1 4 32 0 215 0 ## ## ── Variable type: numeric ────────────────────────────────────────────────────────────────────────────────────────────────────────────────── ## skim_variable n_missing complete_rate mean sd p0 p25 p50 p75 p100 hist ## 1 capture 0 1 421916. 1478638. 0 3280. 33797 221884. 17800000 ▇▁▁▁▁ ## 2 aquaculture 0 1 508368. 4496073. 0 25.2 1574. 25998 63700000 ▇▁▁▁▁ ## 3 total 0 1 930284. 5846301. 0 7270. 44648. 271901. 81500000 ▇▁▁▁▁ ``` ] --- ## Some summary stats ```r fisheries %>% summarise( mean_cap = mean(capture), mean_aqc = mean(aquaculture), mean_tot = mean(total) ) ``` ``` ## # A tibble: 1 x 3 ## mean_cap mean_aqc mean_tot ## <dbl> <dbl> <dbl> ## 1 421916. 508368. 930284. ``` -- <br> .hand[ well, that was boring... ] --- ## A new approach! ```r fisheries %>% summarise(across(capture:total, mean)) ``` ``` ## # A tibble: 1 x 3 ## capture aquaculture total ## <dbl> <dbl> <dbl> ## 1 421916. 508368. 930284. ``` --- .discussion[ The (not-so-great) visualization below shows the distribution of fishery harvest of countries for 2016, by capture and aquaculture. What are some ways you would improve this visualization? Note that countries whose total harvest was less than 100,000 tons are not included in the visualization. ] <img src="img/05/fisheries.png" width="60%" style="display: block; margin: auto;" /> --- class: middle ### Goal: calculate summary statistics at the continent level and visualize them --- ## Data prep .midi[ ```r continents <- read_csv("data/continents.csv") ``` ] -- Filter out countries whose total harvest was less than 100,000 tons since they are not included in the visualization: ```r fisheries <- fisheries %>% filter(total >= 100000) fisheries ``` ``` ## # A tibble: 82 x 4 ## country capture aquaculture total ## <chr> <dbl> <dbl> <dbl> ## 1 Angola 486490 655 487145 ## 2 Argentina 755226 3673 758899 ## 3 Australia 174629 96847 271476 ## 4 Bangladesh 1674770 2203554 3878324 ## 5 Brazil 705000 581230 1286230 ## 6 Cambodia 629950 172500 802450 ## 7 Cameroon 233190 2315 235505 ## 8 Canada 874727 200765 1075492 ## 9 Chad 110000 94 110094 ## 10 Chile 1829238 1050117 2879355 ## # … with 72 more rows ``` --- class: middle, center # Data joins --- .pull-left[ ```r fisheries %>% select(country) ``` ``` ## # A tibble: 82 x 1 ## country ## <chr> ## 1 Angola ## 2 Argentina ## 3 Australia ## 4 Bangladesh ## 5 Brazil ## 6 Cambodia ## 7 Cameroon ## 8 Canada ## 9 Chad ## 10 Chile ## # … with 72 more rows ``` ] .pull-right[ ```r continents ``` ``` ## # A tibble: 245 x 2 ## country continent ## <chr> <chr> ## 1 Afghanistan Asia ## 2 Åland Islands Europe ## 3 Albania Europe ## 4 Algeria Africa ## 5 American Samoa Oceania ## 6 Andorra Europe ## 7 Angola Africa ## 8 Anguilla Americas ## 9 Antigua & Barbuda Americas ## 10 Argentina Americas ## # … with 235 more rows ``` ] --- ## Joining data frames ``` something_join(x, y) ``` Mutating joins: - `inner_join()`: all rows from x where there are matching values in y, return all combination of multiple matches in the case of multiple matches - `left_join()`: all rows from x - `right_join()`: all rows from y - `full_join()`: all rows from both x and y Filtering joins: - `semi_join()`: all rows from x where there are matching values in y, keeping just columns from x. - `anti_join()`: return all rows from x where there are not matching values in y, never duplicate rows of x - ... --- ## Setup For the next few slides... .pull-left[ ```r x ``` ``` ## # A tibble: 3 x 1 ## value ## <dbl> ## 1 1 ## 2 2 ## 3 3 ``` ] .pull-right[ ```r y ``` ``` ## # A tibble: 3 x 1 ## value ## <dbl> ## 1 1 ## 2 2 ## 3 4 ``` ] --- ## `inner_join()` Adds columns to `x` from `y`, matching all rows in `x` AND `y` .pull-left[ ```r inner_join(x, y) ``` ``` ## # A tibble: 2 x 1 ## value ## <dbl> ## 1 1 ## 2 2 ``` ] .pull-right[ <!-- --> ] --- ## `left_join()` Adds columns to `x` from `y`, matching all rows in `x` .pull-left[ ```r left_join(x, y) ``` ``` ## # A tibble: 3 x 1 ## value ## <dbl> ## 1 1 ## 2 2 ## 3 3 ``` ] .pull-right[ <!-- --> ] --- ## `right_join()` Adds columns to `x` from `y`, matching all rows in `y` .pull-left[ ```r right_join(x, y) ``` ``` ## # A tibble: 3 x 1 ## value ## <dbl> ## 1 1 ## 2 2 ## 3 4 ``` ] .pull-right[ 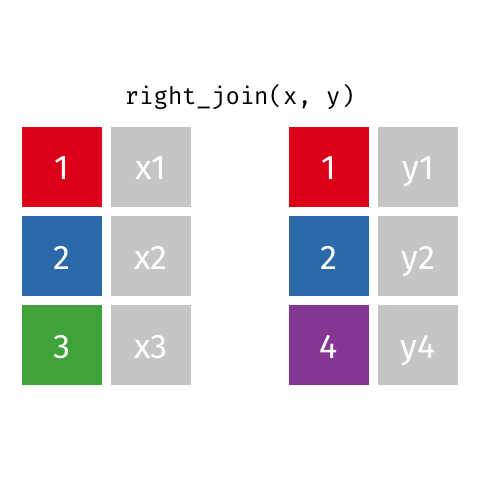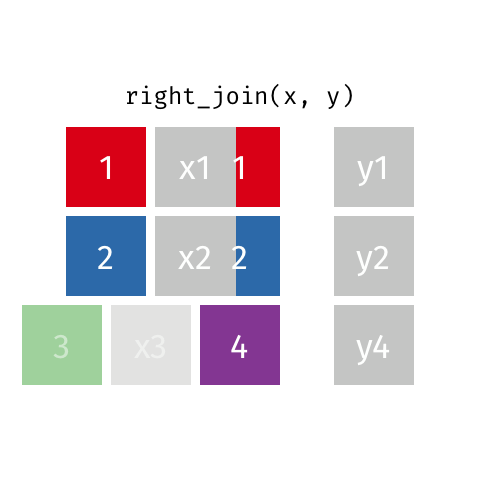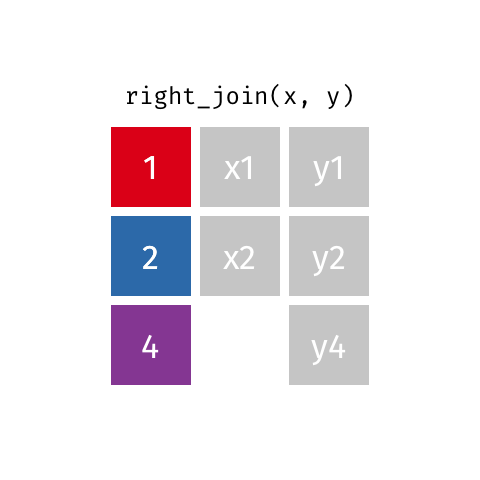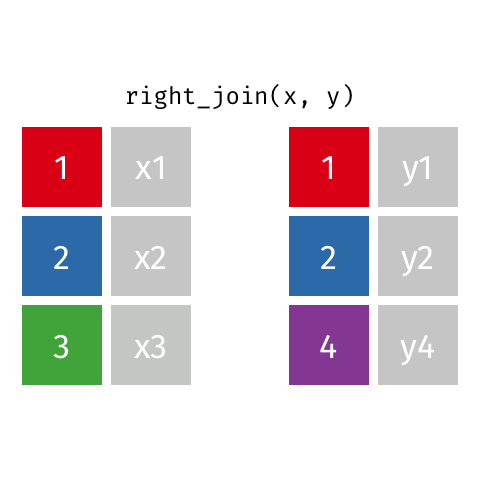<!-- --> ] --- ## `full_join()` Adds columns to `x` from `y`, matching all rows in `x` OR `y` .pull-left[ ```r full_join(x, y) ``` ``` ## # A tibble: 4 x 1 ## value ## <dbl> ## 1 1 ## 2 2 ## 3 3 ## 4 4 ``` ] .pull-right[ 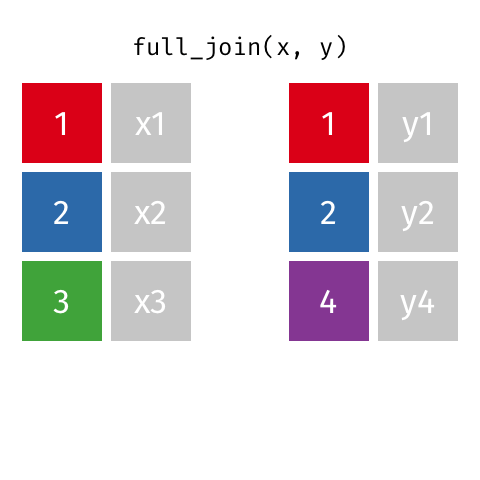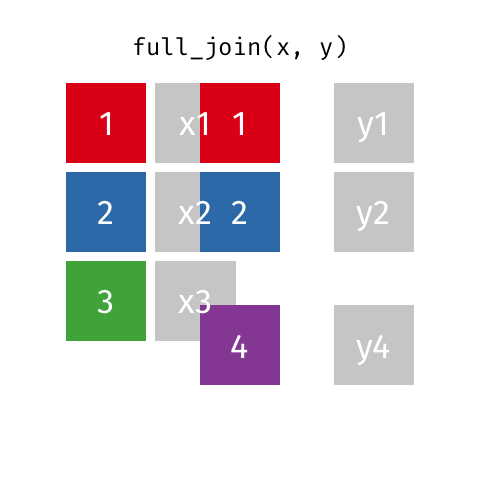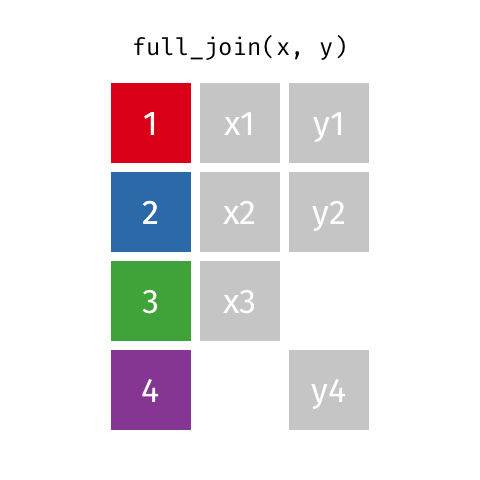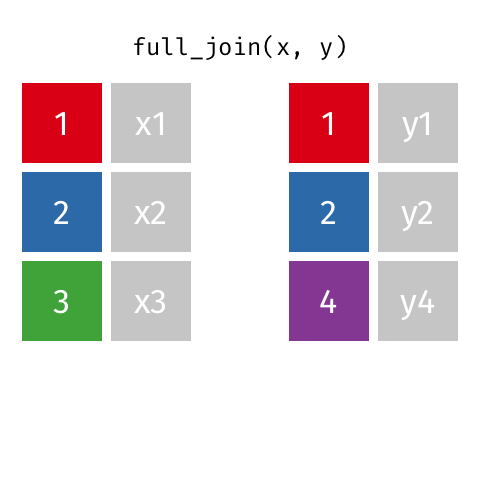<!-- --> ] --- ## `semi_join()` Returns all rows from `x` with a match in `y` (does not add columns from `y`) .pull-left[ ```r semi_join(x, y) ``` ``` ## # A tibble: 2 x 1 ## value ## <dbl> ## 1 1 ## 2 2 ``` ] .pull-right[ <!-- --> ] --- ## `anti_join()` Returns all rows from `x` without a match in `y` (does not add columns from `y`) .pull-left[ ```r anti_join(x, y) ``` ``` ## # A tibble: 1 x 1 ## value ## <dbl> ## 1 3 ``` ] .pull-right[ 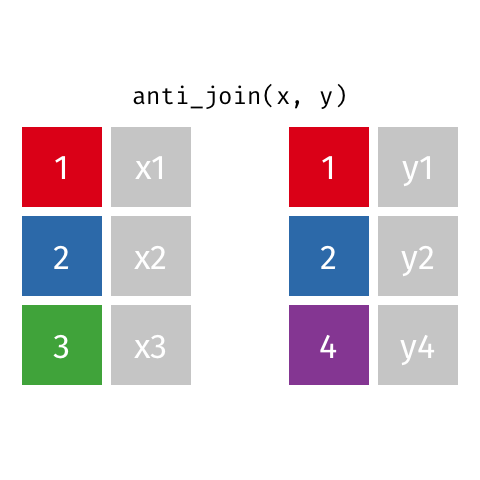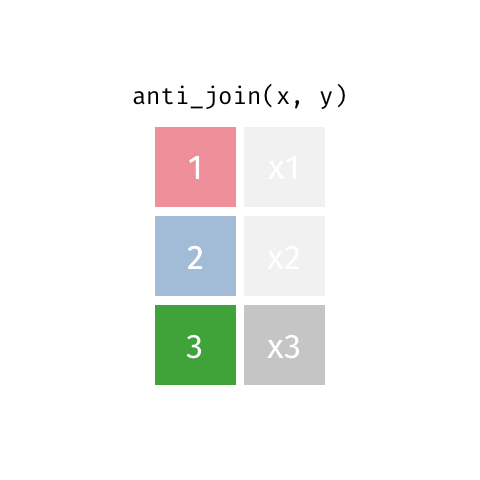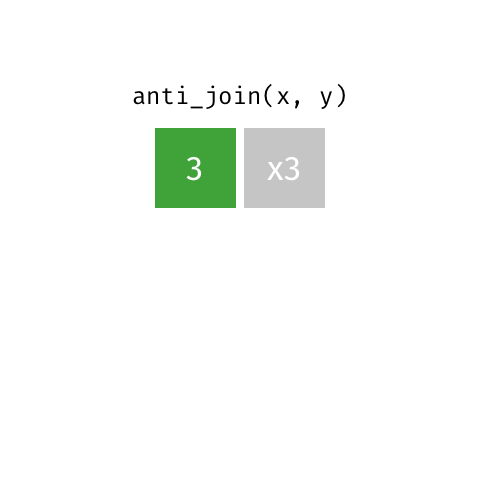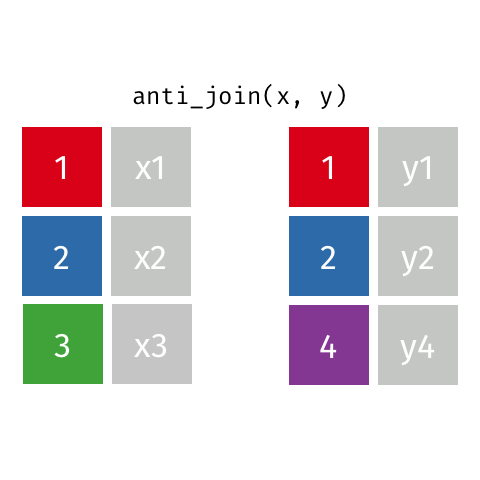<!-- --> ] --- .discussion[ We want to keep all rows and columns from `fisheries` and add a column for corresponding continents. Which join function should we use? ] .pull-left[ ```r fisheries %>% select(country) ``` ``` ## # A tibble: 82 x 1 ## country ## <chr> ## 1 Angola ## 2 Argentina ## 3 Australia ## 4 Bangladesh ## 5 Brazil ## 6 Cambodia ## 7 Cameroon ## 8 Canada ## 9 Chad ## 10 Chile ## # … with 72 more rows ``` ] .pull-right[ ```r continents ``` ``` ## # A tibble: 245 x 2 ## country continent ## <chr> <chr> ## 1 Afghanistan Asia ## 2 Åland Islands Europe ## 3 Albania Europe ## 4 Algeria Africa ## 5 American Samoa Oceania ## 6 Andorra Europe ## 7 Angola Africa ## 8 Anguilla Americas ## 9 Antigua & Barbuda Americas ## 10 Argentina Americas ## # … with 235 more rows ``` ] --- ## Join fisheries and continents ```r fisheries <- left_join(fisheries, continents) ``` -- .discussion[ How does `left_join()` know to join the two data frames by `country`? ] Hint: - Variables in the original fisheries dataset: ``` ## [1] "country" "capture" "aquaculture" "total" ``` - Variables in the continents dataset: ``` ## [1] "country" "continent" ``` --- ## Check the data ```r fisheries %>% slice(11:20) ``` ``` ## # A tibble: 10 x 5 ## country capture aquaculture total continent ## <chr> <dbl> <dbl> <dbl> <chr> ## 1 China 17800000 63700000 81500000 Asia ## 2 Colombia 86344 96970 183314 Americas ## 3 Democratic Republic of the Congo 237372 3161 240533 <NA> ## 4 Denmark 670344 36337 706681 Europe ## 5 Ecuador 715495 451090 1166585 Americas ## 6 Egypt 335614 1370660 1706274 Africa ## 7 Faroe Islands 568435 83300 651735 Europe ## 8 Finland 192065 14412 206477 Europe ## 9 France 561173 166640 727813 Europe ## 10 Germany 271185 41721 312906 Europe ``` --- ## Check the data ```r fisheries %>% filter(is.na(continent)) ``` ``` ## # A tibble: 3 x 5 ## country capture aquaculture total continent ## <chr> <dbl> <dbl> <dbl> <chr> ## 1 Democratic Republic of the Congo 237372 3161 240533 <NA> ## 2 Hong Kong 142775 4258 147033 <NA> ## 3 Myanmar 2072390 1017644 3090034 <NA> ``` --- ## Implement fixes .small[ ```r fisheries <- fisheries %>% mutate(continent = case_when( country == "Democratic Republic of the Congo" ~ "Africa", country == "Hong Kong" ~ "Asia", country == "Myanmar" ~ "Asia", TRUE ~ continent ) ) ``` ...and check again ] ```r fisheries %>% filter(is.na(continent)) ``` ``` ## # A tibble: 0 x 5 ## # … with 5 variables: country <chr>, capture <dbl>, aquaculture <dbl>, total <dbl>, ## # continent <chr> ``` --- .discussion[ What does the following code do? ] ```r fisheries %>% mutate(aquaculture_perc = aquaculture / total) ``` --- class: middle, center ## Demo <!-- .your-turn[ - [RStudio Cloud](http://rstd.io/dsbox-cloud) > `AE 05 - Fisheries + Data joins` > open `fisheries.Rmd` and knit. - Knit the document and work on the Exercise 1. - Once done, place a green sticky on your laptop. If you have questions, place a pink sticky. ] --> --- class: middle, center ## Demo <!-- .your-turn[ - [RStudio Cloud](http://rstd.io/dsbox-cloud) > `AE 05 - Fisheries + Data joins` > open `fisheries.Rmd` and knit. - Knit the document and work on the Exercises 2 - 4. - Once done, place a green sticky on your laptop. If you have questions, place a pink sticky. ] --> --- ## Visualize continent summary stats ```r ggplot(fisheries_summary, aes(x = continent, y = mean_ap)) + geom_col() ``` <!-- --> --- ## Improve visualization ```r ggplot(fisheries_summary, * aes(x = fct_reorder(continent, mean_ap), y = mean_ap)) + geom_col() ``` <!-- --> --- ## Improve visualization further ```r ggplot(fisheries_summary, * aes(y = fct_reorder(continent, mean_ap), x = mean_ap)) + geom_col() + * labs( * x = "", * y = "", * title = "Average share of aquaculture by continent", * subtitle = "out of total fisheries harvest, 2016", * caption = "Source: bit.ly/2VrawTt" * ) + * theme_minimal() ``` See next slide... --- <!-- -->